About
Background
Each organ has a specific function in the body. “Organ-specificity” refers to differential expressions of the same gene across different organs. An organ-specific gene/protein is defined as a gene/protein whose expression is significantly elevated in a specific human organ. An “organ-specific marker” is defined as an organ-specific gene/protein that is also implicated in human diseases related to the organ. Previous studies have shown that identifying specificity for the organ in which a gene or protein is significantly differentially expressed, can lead to discovery of its function. Most currently available resources for organ-specific genes/proteins either allow users to access tissue-specific expression over a limited range of organs, or do not contain disease information such as disease-organ relationship and disease-gene relationship.
Results
We designed an integrated H uman O rgan-specific M olecular E lectronic R epository (HOMER, http://bio.informatics.iupui.edu/homer), defining human organ-specific genes/proteins, based on five criteria: 1) comprehensive organ coverage; 2) gene/protein to disease association; 3) disease-organ association; 4) quantification of organ-specificity; and 5) cross-linking of multiple available data sources. HOMER is a comprehensive database covering about 22,598 proteins, 52 organs, and 4,290 diseases integrated and filtered from organ-specific proteins/genes and disease databases like dbEST, TiSGeD, HPA, CTD, and Disease Ontology. The database has a Web-based user interface that allows users to find organ-specific genes/proteins by gene, protein, organ or disease, to explore the histogram of an organ-specific gene/protein, and to identify disease-related organ-specific genes by browsing the disease data online. Moreover, the quality of the database was validated with comparison to other known databases and two case studies: 1) an association analysis of organ-specific genes with disease and 2) a gene set enrichment analysis of organ-specific gene expression data.
Conclusions
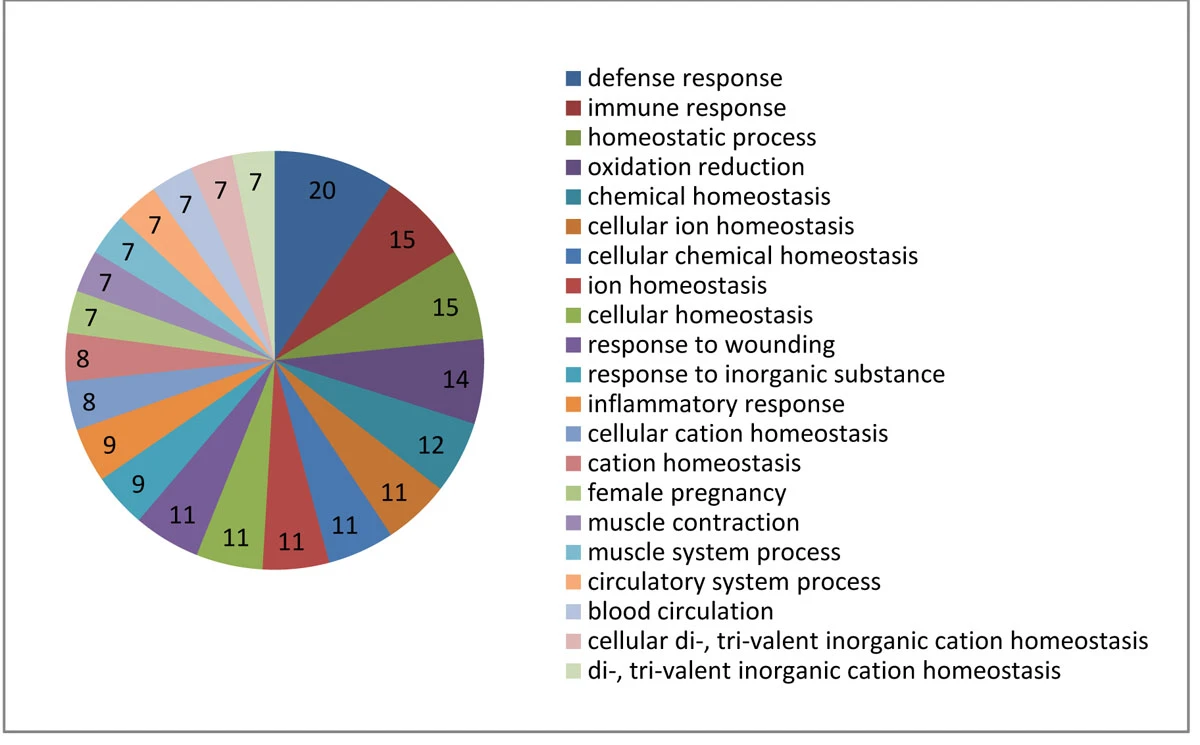
HOMER is a new resource for analyzing, identifying, and characterizing organ-specific molecules in association with disease-organ and disease-gene relationships. The statistical method we developed for organ-specific gene identification can be applied to other organism. The current HOMER database can successfully answer a variety of questions related to organ specificity in human diseases and can help researchers in discovering and characterizing organ-specific genes/proteins with disease relevance.
How to cite us
Zhang, F., Chen, J.Y. HOMER: a human organ-specific molecular electronic repository. BMC Bioinformatics 12, S4 (2011). doi